Cancer Signaling Antibody Array
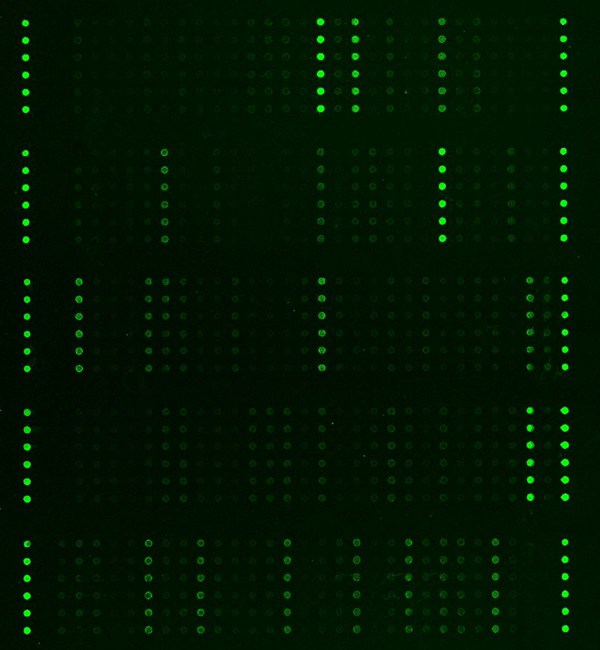
The Cancer Signaling Antibody Array is a high-throughput ELISA based antibody array for qualitative protein phosphorylation profiling, designed for comparing normal samples to treated or diseased samples, and identifying candidate biomarkers. This array features site-specific and phospho-specific antibodies for studying serine, threonine, and tyrosine phosphorylation specific sites. The 269 antibodies on this array allow researchers to examine key proteins from multiple cancer cellular pathways including AKT signaling, apoptosis, EGF/EGFR signaling, ERK/MAPK, p53 signaling and more.
Key Features
- Site-specific phosphorylation profiling and screening with 269 antibodies
- Antibodies covalently immobilized on 3D polymer coated glass slide
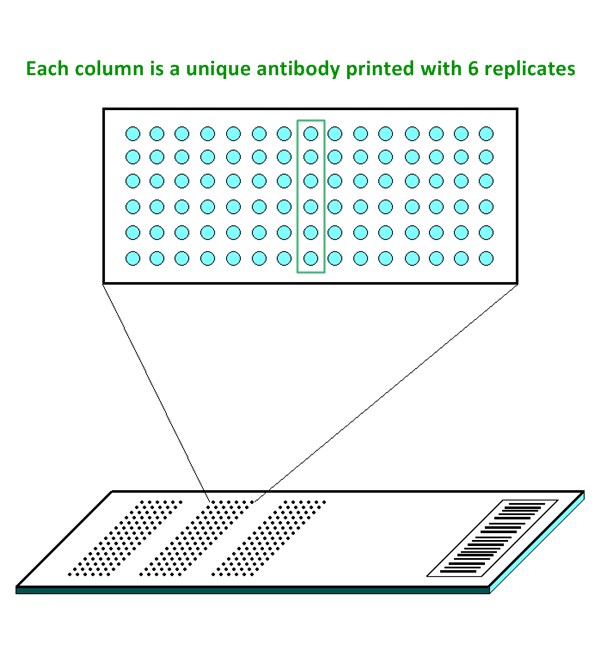
- Six replicates per antibody
- Fluorescent detection
Specifications
| Product Size: | 2 array slides per package for analyzing two samples (untreated vs. treated) |
| Featured Antibodies: | 269 site-specific and phospho-specific antibodies; 6 replicates per antibody |
| Reactivity: | Human: 100% | Mouse: 88% | Rat: 68% |
| Suitable Sample Type: | Cell lysate | Tissue lysate |
| Detection Method: | Fluorescence | Compatible Scanners |
| Internal Controls: | beta-actin | GAPDH | Negative controls |
| Slide dimensions: | 76 x 25 x 1 mm |
| Storage Condition: | 4°C for 6 months |
Product Details
14-3-3 zeta (Ser58), 4E-BP1 (Thr36), AKT (Ser473), AKT (Thr308), AKT2 (Ser474), AMPK1 (Thr174), BAD (Ser112), BAD (Ser136), BAD (Ser155), BCL2 (Ser70), BCL2 (Thr56), BCL-XL (Ser62), BRCA1 (Ser1423), BRCA1 (Ser1524), CaMK2 (Thr286), Catenin b (Ser33), Catenin b (Ser37), Catenin b (Thr41/Ser45), Caspase 3 (Ser150), Caspase 9 (Ser144), Caspase 9 (Thr125), Caspase 9 (Tyr153),Caveolin-1 (Tyr14), CDC2 (Tyr15), CDC25A (Ser75), CDC25C (Ser216), CDK2 (Thr160), Chk1 (Ser280), Chk1 (Ser317), Chk1 (Ser345), Chk2 (Ser516), Chk2 (Thr68), c-Jun (Thr239), c-Jun (Ser243), c-Jun (Ser73), c-Kit (Tyr721), CREB (Ser133), CrkII (Tyr221), eEF2K (Ser366), EGFR (Tyr1110), eIF2A (Ser51), eIF4E (Ser209), Elk1 (Ser383), ERK3 (Ser189), ERK8 (Thr175/Tyr177), Estrogen Receptor-a (Ser167), FAK (Tyr397), FAK (Tyr861), FAK (Tyr925), FGFR1 (Tyr154), FKHR (Ser256), GSK3 alpha (Ser21), GSK3 beta (Ser9), HDAC8 (Ser39), HER2 (Tyr877), Histone H2A.X (Ser139), HSF1 (Ser303), HSP27 (Ser15), HSP27 (Ser78), HSP90B (Ser254), ICAM-1 (Tyr512), IGF-1R (Tyr1161), IkB-alpha (Ser32/36), IkB-alpha (Tyr42), IkB-beta (Ser23), IkB-epsilon (Ser22), IKK-alpha (Thr23), Integrin beta-3 (Tyr773), Integrin beta-3 (Tyr785), JAK1 (Tyr1022), JAK2 (Tyr1007), JAK2 (Tyr221), JunB (Ser259), JunB (Ser79), JunD (Ser255), Keratin 18 (Ser33), MEK1 (Ser217), MEK1 (Ser221), MEK1 (Thr291), MEK-2 (Thr394), Met (Tyr1349), MKK3 (Ser189), MSK1 (Ser376), mTOR (Ser2448), Myc (Ser62), Myc (Ser373), Myc (Thr358), Myc (Thr58), NFkB-p100/p52 (Ser865), NFkB-p100/p52 (Ser869), NFkB-p105/p50 (Ser893), NFkB-p105/p50 (Ser907), NFkB-p105/p50 (Ser932), NFkB-p65 (Ser529), NFkB-p65 (Thr254), p21Cip1 (Thr145), p27Kip1 (Ser10), p27Kip1 (Thr187), P38 MAPK (Thr180), P38 MAPK (Tyr182), p44/42 MAPK (Thr202), p44/42 MAPK (Tyr204), p53 (Ser6), p53 (Ser315), P70S6K (Ser424), PDGF R beta (Tyr751), PDK1 (Ser241), PI3K p85a (Tyr607), PTEN (Ser380/Thr382/Thr383), Pyk2 (Tyr402), Rac1/cdc42 (Ser71), Raf1 (Ser259), Rb (Ser780), Rel (Ser503), SAPK/JNK (Thr183), Shc (Tyr349), SHP-2 (Tyr580), Smad3 (Ser425), Src (Tyr418), Src (Tyr529), STAT1 (Ser727), STAT1 (Tyr701), STAT3 (Ser727), STAT3 (Tyr705), STAT4 (Tyr693), STAT5A (Ser780), STAT5A (Tyr694), STAT6 (Thr645), STAT6 (Tyr641), Tau (Ser404), Trk B (Tyr515), TYK2 (Tyr1054), VEGFR2 (Tyr951)
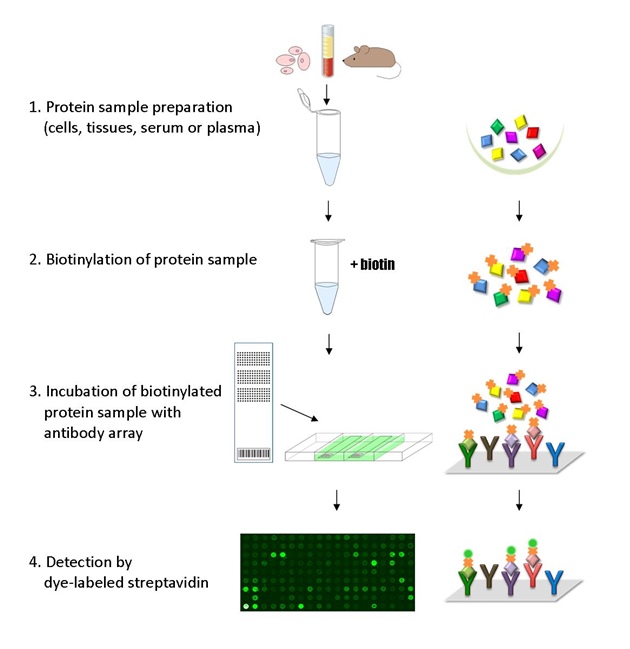
The ELISA based Cancer Signaling Phospho Antibody Array platform involves four major steps:
- Protein extraction with non-denaturing lysis buffer
- Biotinylation of protein samples
- Incubation of labeled samples with antibody array
- Detection by dye conjugated streptavidin
![]() GAL File (To download, right click on the file name, then choose “Save target as”)
GAL File (To download, right click on the file name, then choose “Save target as”)
Dai B, Shi X, HMQ‐T‐B10 induces human liver cell apoptosis by competitively targeting EphrinB2 and regulating its pathway, J Cell Mol Med, 2018, 22(11): 5231–5243
Gu C, Liu Y, Discovery of the oncogenic parp1, a target of bcr-abl and a potential therapeutics, in identification of mir-181a/PPFIA1 signaling pathway, Mol Ther Nucleic Acids, 2019 Feb 8, https://doi.org/10.1016/j.omtn.2019.01.015
El-Haibi CP, Singh R, Antibody Microarray Analysis of Signaling Networks Regulated by Cxcl13 and Cxcr5 in Prostate Cancer, Journal of Proteomics and Bioinformatics 2012, 5(8): 177-184
Koncar RF, Dey BR, Identification of novel RAS signaling therapeutic vulnerabilities in Diffuse Intrinsic Pontine Gliomas, Cancer Res. 2019 Jun 14. doi: 10.1158/0008-5472.CAN-18-3521
Liu D, Zhang XX, C/EBPβ enhances platinum resistance of ovarian cancer cells by reprogramming H3K79 methylation, Nat Commun 2018, 9(1):1739. doi: 10.1038/s41467-018-03590-5
Mathison A, Kerketta R, Kras-G12D induces changes in chromatin territories that differentially impact early nuclear reprogramming in pancreatic cells. Genome Biol 2021; 22:289. https://doi.org/10.1186/s13059-021-02498-6
Nekova T, Kneitz S, Silencing of CDK2, but not CDK1, separates mitogenic from anti-apoptotic signaling, sensitizing p53 defective cells for synthetic lethality, Cell Cycle 2016 (23):3203-3209
Oda K, Umemura M, Transient receptor potential cation 3 channel regulates melanoma proliferation and migration, J Physiol Sci, 2017; 67(4):497-505
Ward A, Mir H, Quercetin inhibits prostate cancer by attenuating cell survival and inhibiting anti-apoptotic pathways, World J Surg Oncol. 2018 Jun 14;16(1):108
Watanabe T, Takahashi A, Epithelial–mesenchymal transition in human gastric cancer cell lines induced by TNF-α-inducing protein of Helicobacter pylori, International Journal of Cancer 2014 134(10):2373-82. doi: 10.1002/ijc.28582
Xia Z, Huang M, Cadherin Related Family Member 2 Acts As A Tumor Suppressor By Inactivating AKT In Human Hepatocellular Carcinoma, J Cancer 2019, 10(4): 864-873
Xu Y, Dong B, Sphingosine kinase 1 overexpression contributes to sunitinib resistance in clear cell renal cell carcinoma, OncoImmunology, 2018 Sep 25;7(12):e1502130
Zhang J, Gelman IH, Glucose drives growth factor-independent esophageal cancer proliferation via phosphohistidine-FAK signaling, Cell Mol Gastroenterol Hepatol. 2019 Mar 2. pii: S2352-345X(19)30025-6. doi: 10.1016/j.jcmgh.2019.02.009
Services
If you don’t have access to a microarray, send the finished arrays to our lab for scanning. Raw scan images are delivered in tiff format.
Cost: Free
Array Image Quantification and Analysis Service includes data extraction, data organization and analysis of the array images obtained through our array scanning service.
Cost: $260 per slide
Complete Antibody Array Assay Service allows investigators to send research samples to our laboratory for analysis. There is no need to purchase the arrays and reagents and running the assays yourself. Simply select the array of your choice, and then send off the samples to our lab. This convenient hands-off approach offers quick turnaround and reliable results, saving you valuable time and resources. All assays will be performed by our highly trained scientists at our headquarter in Sunnyvale, California. Results are delivered by email in 1-3 weeks.
Cost: $1,565 per sample